| |
Contents | EBJ Home | Single molecule measurements and biological motors |
 |
||
| Cy3-EDA-ATP binding to myosin | ||
As mentioned previously, single molecule fluorescence can be used to measure kinetic parameters. Here we give an example of how this can be done. In general, kinetic parameters measured by this method should be close to those measured by conventional techniques, but one must be wary of a number of potential artefacts. For example, when measuring dwell times it is important to be sure that one is measuring the tue dwell time and not the photobleaching of your probe. It is also important to be aware that simply attaching the proteins being studied to a surface may change their behaviour.
In the example shown here, we used a fluorescent ATP analogue, Cy3-EDA-ATP, to measure the ADP release rate of a myosin I immobilised on a quartz surface. In this myosin, ADP release is the rate limiting step, so measuring the dwell time of the spots provides an estimate of the first order rate constant for ADP release. The first figure shows a false colour animation of the fluorescence. The video data was digitised and each pixel was median filtered with a 3 frame window to reduce intensifier noise. The dwell time of each spot was then measured off from a time series trace. The dwell times were accumulated in 1 s bins and an exponential was fitted to the data. The rate constant obtained (0.16 s-1) was consistent with that measured by bulk techniques (0.18s-1, Jontes et al., 1997).
Single molecules of fluorescent ATP binding to and dissociating from myosin were first observed by Funatsu et al. (1995). This technique was combined with optical tweezers by Ishijima et al. (1998) to investigate mechanochemical coupling in myosin.
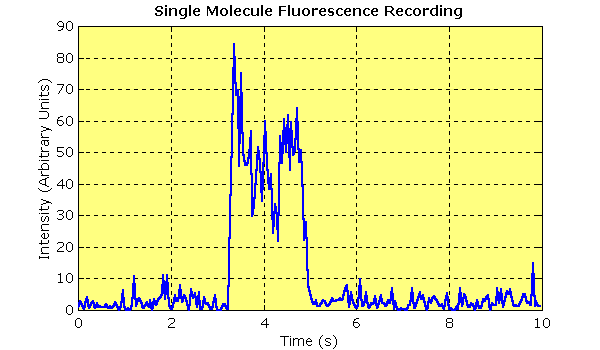 |
| Single molecule binding event. This plot shows 10 seconds of unfiltered data from a single molecule fluorescence experiment. TIRF was used to visualise the binding of a fluorescent nucleotide analogue (10 nM Cy3-EDA-ATP) to a quartz surface sparsely coated with myosin I. The data were recorded using an image-intensified video camera and digitised with a frame grabber. The trace shows the intensity of the spot over time (integrated over a 3 × 3 pixel area). Intensity rises and falls in a single step as nucleotide arrives and departs. The durations of these events were measured and plotted as a histogram. |
| Contents | Next |